AusTrakka surpasses 200,000 sequences of SARS-CoV-2
Find out more about the significance of this milestone
New genomics insights strengthen foodborne disease tracking across Australasia
AusPathoGen shared new national insights on Salmonella and Listeria surveillance at the annual Australian/New Zealand Enteric Reference Network (ANZERN) meeting on November 7th, highlighting progress toward harmonised genomic methodologies and faster, decentralised systems for enteric disease monitoring.
Collaboration through ANZERN
Members of the Australia and New Zealand Enterics Reference Network (ANZERN) met in Adelaide for their annual face-to-face meeting, hosted by SA Pathology. The network connects Enteric Pathogens sections across Public Health Reference Laboratories, fostering collaboration and alignment across both countries.
AusPathoGen was invited to present updates on national genomics projects for Salmonella and Listeria, and represented by AusPathoGen Senior Research Fellow Dr Patiyan Andersson. Both projects demonstrate how genomic data is transforming surveillance, improving consistency, and strengthening public health responses.
Advancing national Salmonella genomics
Dr Andersson shared findings from the AusPathoGen non-typhoidal Salmonella project — a national cross-sectional analysis designed to inform harmonisation of national analysis methodologies and implementation of genomic surveillance strategies.

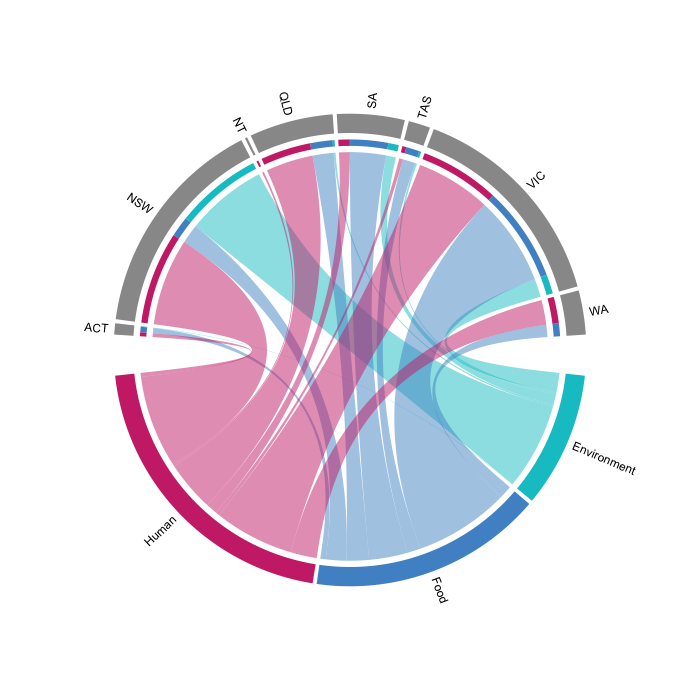
The project has collated 18,200 genomic sequences from two national snapshots (January–March 2023 and 2024) during which all notified samples were attempted to be sequenced, in addition to retrospective data from 2018–2022. This comprehensive dataset is helping to identify priority serovars for genomic analysis for optimal resource utilisation and establish national methodologies for screening to identify sample groups for investigation and efficient high resolution analysis suitable for automation — laying the foundation for a consistent, nationwide approach to genomics-based Salmonella surveillance.
“This work is a crucial step toward nationally coordinated, genomics-driven surveillance of enteric pathogens.” - Dr. Patiyan Andersson, AusPathoGen Senior Research Fellow
The second presentation focused on the national Listeria genomics surveillance system and the impact of decentralising sequencing across jurisdictions.
Since routine genomic sequencing began in 2016, capacity has expanded beyond the National Listeria Reference Laboratory in Melbourne to include four jurisdictions undertaking local sequencing. A review of 1,200 samples (2016–2023) showed a statistically significant reduction in median overall turnaround times from 32 days to 26 days during the genomics era.
Although turnaround times initially increased during transitions to local sequencing, they improved significantly as workflows and referral processes were established. The project provided tangible evidence of the benefits of democratising sequencing capacity across public health laboratories. Findings were published in Emerging Infectious Diseases (2025): https://pubmed.ncbi.nlm.nih.gov/40359100/
Building on the work to-date, AusPathoGen will in its continuation consolidate national capability in pathogen genomics through a federated data sharing and analysis platform, embedding genomics within Australia’s national communicable disease surveillance infrastructure.
Future activities include:
- Implement national genomic surveillance and data sharing
- Enhance preparedness and response to emerging disease threats
- Expand integration across One Health and clinical domains
- Embed evaluation and workforce development to ensure sustainability
By establishing genomics as a core component of Australia’s infectious disease surveillance and response system, AusPathoGen continues to drive collaboration, innovation, and national coordination in public health genomics.